WARNING! Dysbiosis Induced By SARS-CoV-2 Infections Presents Risk Of Antibiotic Resistance!
Researchers from Capital Medical University-China, Hong Kong University of Science and Technology-Hong Kong SAR, Tsinghua University-China and Peking University-China have in a new study found that altered microbiomes in SARS-CoV-2 infection pose threat of antibiotic resistance.
Microbiome plays a critical role in modulating the human host metabolism and immune system. Connections and interactions have been found between the microbiome of the gut and oral-pharynx in the context of SARS-CoV-2 and other viral infections, hence, to broaden the understanding of host-viral responses in general and to deepen the knowledge of COVID-19, the study team performed a large-scale, systematic evaluation of the effect of SARS-CoV-2 infection on human microbiota in patients with varying disease severity.
The study team processed 521 samples from 203 COVID-19 patients with varying disease severity and 94 samples from 31 healthy donors, consisting of 213 pharyngeal swabs, 250 sputum, and 152 faecal samples, and obtained meta-transcriptomes as well as SARS-CoV-2 sequences from each sample.
The study findings from detailed assessment of these samples revealed altered microbial composition and function in the upper respiratory tract (URT) and gut of COVID-19 patients, and these changes are significantly associated with disease severity.
Interestingly, the upper respiratory tract (URT) and gut microbiota show different patterns of alteration, where gut microbiome seems to be more variable and in direct correlation with viral load; and microbial community in upper respiratory tract renders high risk of antibiotic resistance. Longitudinally, microbial composition remains relatively stable during the study period.
The study findings revealed different trends and the relative sensitivity of microbiome in different body sites to SARS-CoV-2 infection.
Also, although the use of antibiotics is often essential for prevention and treatment of secondary infections, the study findings indicate a need to evaluate potential antibiotic resistance in the management of COVID-19 patients in the ongoing pandemic. Moreover, longitudinal follow-up to monitor the restoration of the microbiome could enhance the understanding of the long-term effects of COVID-19.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.researchsquare.com/article/rs-1371786/v1
The COVID-19 pandemic has caused severe disruption of the health, social and economic systems around the world for almost 25 months. Despite the rapid development and rollout of vaccines against the causative severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the subsequent emergence of a number of variants of concern (VOCs) has challenged the ability to control the spread of SARS-CoV-2.
It remains important to understand how SARS-CoV-2 causes disease and how its treatment affects humans.
This study by researchers from China discusses changes in the microbiota of various parts of the body with SARS-CoV-2 infection.
The SARS-CoV-2 coronavirus binds the host cell angiotensin-converting enzyme 2 (ACE2) to subsequently enter cells to cause productive infection. The major target of SARS-CoV-2 is the respiratory tract, and COVID-19 pneumonia is a frequent occurrence.
/>
But it should also be noted that many other organs are also known to be involved in SARS-CoV-2 infection, as ACE2 receptors are present in multiple types of organs and tissues. In fact, SARS-CoV-2 particles and nucleic acids have been identified in bronchoalveolar lavage fluid (BALF) samples, sputum, swabs from the nose and throat, blood, feces, and saliva.
Corresponding author, Dr Angela Ruohao Wu from Hong Kong University of Science and Technology told Thailand Medical News, “Importantly it is already known that respiratory viruses can disrupt the microbial community on the mucosa of the gut and respiratory tract. This can trigger secondary bacterial infection, thereby increasing the severity of the illness and worsening the outcome.”
In order to further explore this, the study team used a ribonucleic acid sequencing (RNA-seq) library construction strategy called MINERVA, which accelerates the time required for sequencing SARS-CoV-2 and metatranscriptomics from different parts of the body in COVID-19 patients.
Importantly the types of changing patterns in the gut were compared to those in the respiratory tract in order to elucidate their relationship with SARS-CoV-2 load.
The research included almost 540 samples obtained from over 200 COVID-19 patients. All samples were taken within two weeks of symptom onset.
The study team found that respiratory tract species were less diverse in patients as compared to controls, though species were equally rich in all pharyngeal, feces, and sputum samples. Despite the absence of any obvious difference in alpha diversity in the gut microbiome, there is a definite change in the flora in these patients.
Significantly, the most severe illness was linked to the greatest reduction in diversity in all types of samples. With COVID-19, respiratory samples showed a reliable reduction in Shannon diversity; however, in feces samples, though the alpha diversity remained unchanged, the microbial composition was altered. This was due to an increase in SARS-CoV-2, along with antibiotic and antiviral therapeutics.
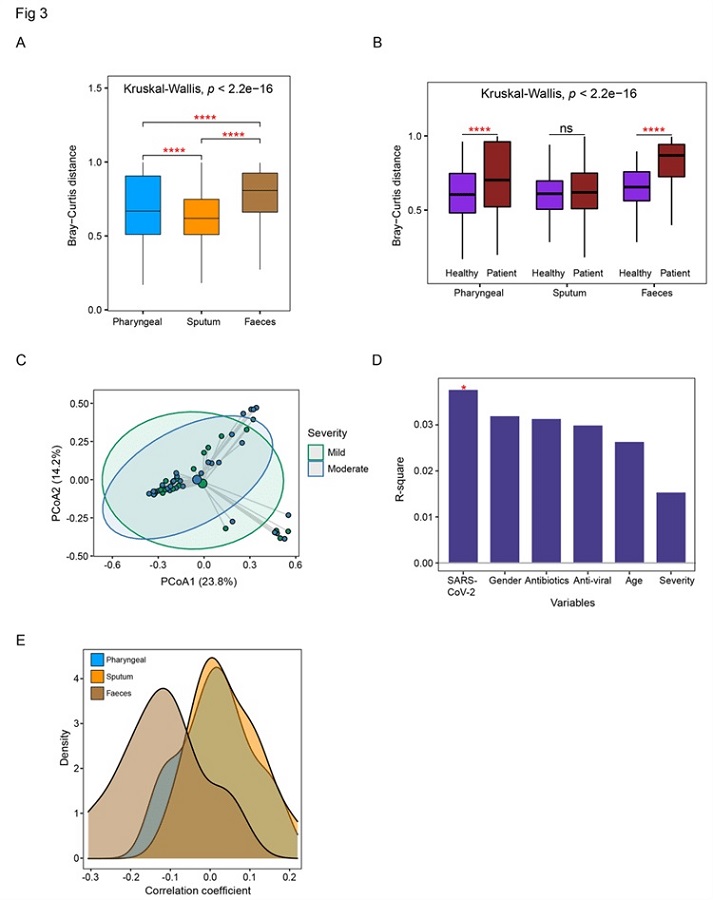 Detailed comparison of the dysbiosis patterns of microbial composition in respiratory tract and gut samples. (A) Comparison of Bray-Curtis distance between patients and healthy controls in three sample types. (B) Comparison of the within-patient and within-healthy control Bray-Cutis distance in three sample types. (C) PCoA analysis of the microbial composition in faeces samples from mild and moderate patients. (D) PERMANOVA test to identify meta factors potentially associated with the microbial composition in faeces samples from mild and moderate patients. (E) Distribution of the correlation coefficients between SARS-CoV-2 and representative species in three sample types. Kruskal-Wallis test was used for multiple-group comparison and Wilcox rank-sum test was used for post-hoc two-group comparison.
Detailed comparison of the dysbiosis patterns of microbial composition in respiratory tract and gut samples. (A) Comparison of Bray-Curtis distance between patients and healthy controls in three sample types. (B) Comparison of the within-patient and within-healthy control Bray-Cutis distance in three sample types. (C) PCoA analysis of the microbial composition in faeces samples from mild and moderate patients. (D) PERMANOVA test to identify meta factors potentially associated with the microbial composition in faeces samples from mild and moderate patients. (E) Distribution of the correlation coefficients between SARS-CoV-2 and representative species in three sample types. Kruskal-Wallis test was used for multiple-group comparison and Wilcox rank-sum test was used for post-hoc two-group comparison.
Interestingly, the greatest increase in microbial abundance was in Streptococcus that was present in respiratory samples, both normal and opportunistic species. In fecal samples, there was no difference in the most abundant species, of which included Bacteroides. However, Fecalibacterium showed a reduction, whereas Acinetobacter, Citrobacter, and Pseudomonas increased in feces samples.
Importantly and also worryingly, many of the species that were reduced were involved in the production of short-chain fatty acids, engaged in the regulation of gut health, and participated in the gut inflammatory response.
Also, certain species that showed increased numbers in COVID-19 were not typical of human feces. One example is Pseudomonas fluorescens, which is a low pathogenic bacterium, and Halomonas, which is usually an inhabitant of marine environments that is usually a commensal organism that is sometimes responsible for hospital infections.
The gut microbiota was more severely affected by SARS-CoV-2 infection as compared to respiratory samples, thus demonstrating the impact of disease severity and antibiotic treatment. Overall, the composition of the microbiota remained relatively stable during the study period.
Interestingly, the dysbiosis profile in the gut varied in functional alterations from that in the respiratory tract. In the gut, carbohydrate metabolism was affected along with single-chain fatty acid (SCFA) production, with a higher expression of genes related to the stress response. Conversely, stress genes and toxin genes predominated in the respiratory tract microbiome.
Also worrying, in pharyngeal samples, the transportation of many different molecules appeared to be a focus of gene upregulation in patient samples, as demonstrated by the higher level of expression of iron, zinc, manganese, or copper transport molecules. Cellular components that may indicate a higher risk of antibiotic resistance, such as multidrug efflux pump SatA and SatB, were enriched in severe COVID-19. Notably, both SatA and SatB participate in norfloxacin and ciprofloxacin ABC transport.
It was also found that in the sputum, transport genes, stress response genes, and virulence genes were similarly abundant in patients as compared to healthy controls.
Numerous genes in the respiratory tract microbiome also focused on the stress response, thereby indicating that the microbes were experiencing stress, whether due to SARS-CoV-2 infection or treatment.
The fecal samples in healthy controls showed more genes responsible for fatty acid metabolism, including SCFA. However, in COVID-19 patients, genes expressed at higher levels were primarily responsible for amino acid metabolism and the stress response.
The study findings show that with COVID-19, and depending on the severity of illness, different patterns of alterations in the microbiomes of the gut and upper respiratory tract are observed. Both the composition and function of the microbiomes of these regions were affected.
The upper respiratory tract samples showed less diversity of the microbiome, perhaps because normal species were lost and Streptococcus species expanded.
Importantly, in the gut samples, beneficial species that generate SCFA were lost. This activity is key to gut mucosal integrity, regulation of metabolism, and homeostasis of the immune system.
Furthermore, along with the depletion of normal species in both the gut and respiratory microbiomes, gut bacteria responded more readily to the abundance of SARS-CoV-2 and showed a wider range of responses among patients. This included loss of carbohydrate and fatty acid metabolism, including the production of SCFA.
The functional changes in the respiratory microbiome included a major increase in stress response and toxin response genes. These changes, which affect microbiome homeostasis, could persist for months after recovery, according to earlier research. https://pubmed.ncbi.nlm.nih.gov/35082169/
The microbiome changes observed in this study support the evolution of antibiotic resistance during the phase of acute illness. This could prolong the recovery of the normal microflora at each location and cause delayed effects of the viral illness.
Although the prophylactic application of antibiotics is sometimes essential for prevention and treatment of secondary infections, close monitoring and strategies for more precise use of antibiotics is urgently required in the ongoing SARS-CoV-2 pandemic.
Please help support this website by making a donation to assists in sustaining this website and to help support all our research and community initiatives. Your help saves lives directly and indirectly. https://www.thailandmedical.news/p/sponsorship
For the latest on SARS-CoV-2 and Microbiome, keep on logging to Thailand Medical News.

 Detailed comparison of the dysbiosis patterns of microbial composition in respiratory tract and gut samples. (A) Comparison of Bray-Curtis distance between patients and healthy controls in three sample types. (B) Comparison of the within-patient and within-healthy control Bray-Cutis distance in three sample types. (C) PCoA analysis of the microbial composition in faeces samples from mild and moderate patients. (D) PERMANOVA test to identify meta factors potentially associated with the microbial composition in faeces samples from mild and moderate patients. (E) Distribution of the correlation coefficients between SARS-CoV-2 and representative species in three sample types. Kruskal-Wallis test was used for multiple-group comparison and Wilcox rank-sum test was used for post-hoc two-group comparison.
Detailed comparison of the dysbiosis patterns of microbial composition in respiratory tract and gut samples. (A) Comparison of Bray-Curtis distance between patients and healthy controls in three sample types. (B) Comparison of the within-patient and within-healthy control Bray-Cutis distance in three sample types. (C) PCoA analysis of the microbial composition in faeces samples from mild and moderate patients. (D) PERMANOVA test to identify meta factors potentially associated with the microbial composition in faeces samples from mild and moderate patients. (E) Distribution of the correlation coefficients between SARS-CoV-2 and representative species in three sample types. Kruskal-Wallis test was used for multiple-group comparison and Wilcox rank-sum test was used for post-hoc two-group comparison.